Research
How does non-trivial collective behavior of a complex system emerge from the interactions among its components? This question motivated my research on vortices in superconductors, on stability of emulsions in liquid crystals, and, more recently, on evolutionary dynamics. Evolution is a particularly rich example of emergent complex behavior. By studying evolutionary dynamics, we can understand not only the origin and development of life, but also some general features of nonequilibrium processes. A quantitative understanding of evolution and ecology is also necessary to fight cancer and HIV, create a sustainable agriculture, or prevent the spread of antibiotic resistance. My ambition is to understand how various evolutionary and ecological forces prevent or enable adaptation. I am especially interested in connecting my work to the growing number of evolutionary experiments with microbes and the increasing amount of biomedical data. The former enables rigorous tests of the existing theories, while the latter is an exciting opportunity to both learn about evolution and contribute to the improvement of the human condition.
A few highlights from my work are described below.
Motivation
Spatial expansions are a recurrent theme in ecology and evolution. They describe the spread of invasive species, epidemics, and tumor growth among many other phenomena ranging from cellular to continental scales. Since expanding populations often need to adapt to the new environment, there is often a very strong coupling between ecological and evolutionary dynamics. The outcome of this coupling is seemingly unpredictable, which complicates the control of often undesirable growth. While invasion velocity rapidly increase in some species, other species accumulate deleterious mutations and slow down. One explanation for such striking differences is demographic fluctuations (genetic drift), which control the efficacy of natural selection. Deleterious mutations can stochastically reach fixation in populations with strong genetic drift, but only beneficial mutations fix when the genetic drift is weak.
Quantifying genetic drift is an old and well-understood problem in well mixed populations, but it is much more difficult in expanding populations. Indeed, demographic fluctuations are very strong at the leading edge of the expansion front, but these fluctuations are counteracted by constant reseeding from the population bulk. The mathematical formulation of these dynamics leads to nonlinear stochastic partial differential equations that have attracted sustained interest in mathematics and statistical physics because of their simple formulation and yet very rich and hard to understand dynamics. Furthermore, these equations describe many important phenomena from crystal growth to scattering problems in quantum chromodynamics.
Prior work focused mostly on the expansion velocity and established that all moving fronts can be classifies as either “pulled” or “pushed” depending on who wins the competition between the growth at the edge and reseeding from the bulk. The velocity of pulled expansions depends only on the growth rate at low population densities, i.e. the leading edge effectively pulls the expansions. In contrast, the velocity of pushed expansions exceeds the value that one would expect from the growth rate at the leading edge. Thus, the expansion is effectively pushed from the regions of higher growth rate behind the front. The analysis of fixation probabilities further showed that successful mutations occur only at the leading edge in pulled waves, but are spread throughout the expansion from in pushed waves. Given this, one naturally expect that genetic drift is higher in pulled than in pushed waves, but little was known beyond this expectation.
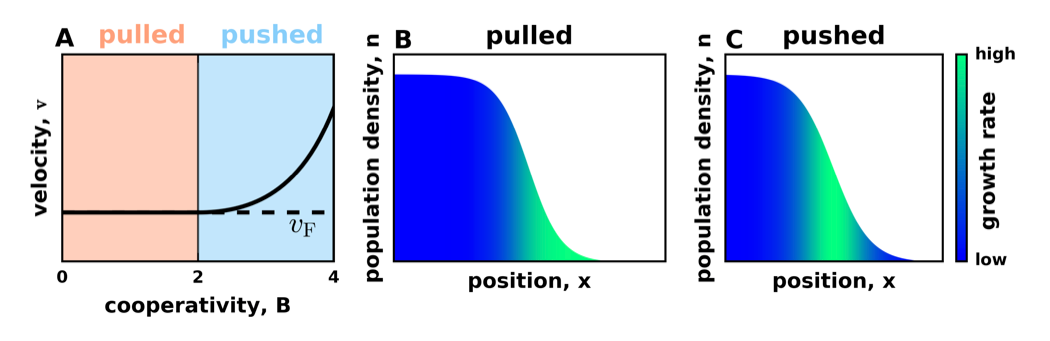 A illustrates that the velocity of pulled waves is given by the Fisher-Kolmogorov velocity, but the velocity is higher than this prediction for pushed waves. B and C illustrate the distribution of growth rate within the expansion front.
A illustrates that the velocity of pulled waves is given by the Fisher-Kolmogorov velocity, but the velocity is higher than this prediction for pushed waves. B and C illustrate the distribution of growth rate within the expansion front.
A new class of waves: neither pulled nor pushed
We systematically explored how genetic drift changes at the transition from pulled to pushed expansions. Surprisingly, we found not two, but three universality classes with very different pattern of fluctuations. For example, genetic diversity scales logarithmically, as a power law, or linearly with carrying capacity in each of the classes. Similar scalings apply to the effective diffusion constant of the front and other metrics of fluctuations. We computed all scaling exponents analytically and showed that they depend on a single parameter: the ratio of the expansion velocity to the geometric mean of dispersal and growth rates at the leading edge. More recently, we have also shown that the same theory holds for density-dependent dispersal. In such circumstances, expansions are pushed because the dispersal rates peak at higher population densities behind the leading edge.
Expansions create commonly observed, yet often unexplained genealogical structures.
We found that evolutionary outcomes could be very sensitive to the details of the dispersal and growth. A few percent change in the velocity ratio defined above could lead to orders of magnitude difference in genetic diversity. Thus, populations could have dramatically different potential to evolve drug resistance or adapt to other challenges. Furthermore, our work suggests that large variations in the genetic diversity of invasive species could be attributed to subtle density-dependence in their growth and dispersal rates, which in most cases have not been carefully characterized. We also predict that the genealogies of expanding populations will be markedly different between the universality classes. In particular, we expect that the classic Kingman coalescent describes only one of the universality classes while the other two have genealogies with multiple merges that are described by the Lambda coalescent. Numerical work to test this prediction is ongoing.
For more details see the following papers:
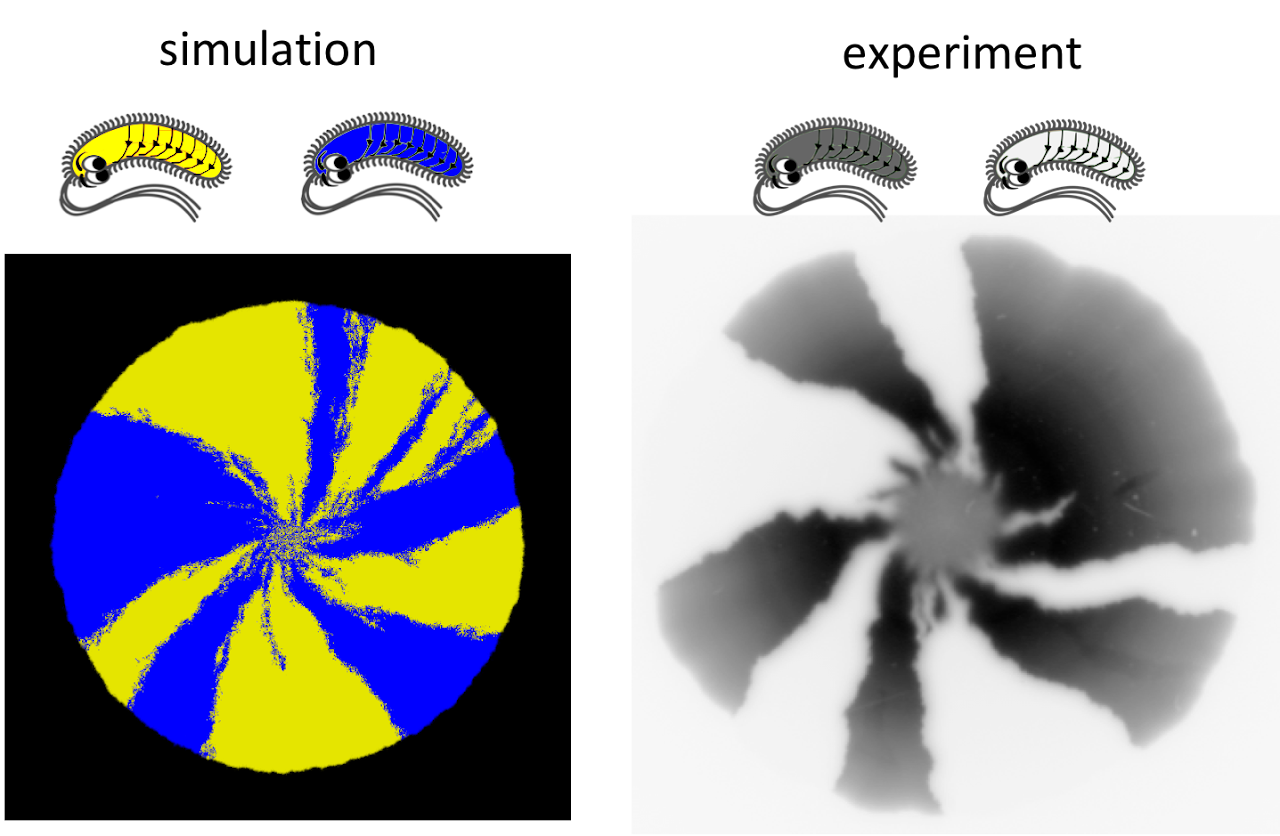 Fig. A: Spiraling motion of strain boundaries reveal chiral patterns of growth. Strains are identical except for the color. Segregation into sectors occurs due to genetic drift.
Fig. A: Spiraling motion of strain boundaries reveal chiral patterns of growth. Strains are identical except for the color. Segregation into sectors occurs due to genetic drift.
We found that chirality affects the competition between the strains by deforming the shape of the cellular aggregate. Deformations occur at the boundaries between the strains and come in two flavors. At half of the boundaries, the strains move towards each other, which results in the formation of bulge (Fig. D). Dips form at the other half of the boundaries, where the strains move away from each other. Bulges grow much faster than the dips, and, eventually, completely fill the edge of the aggregate. The relative abundance of the strains is then determined by the slopes of the bulged, which are controlled by the relative chirality of the strains.
The dynamics described above are fully captured by a simple analytical theory that includes two equations: one for the shape of the aggregate and one for the relative abundance of the strains. These equations are KPZ and Burger’s equations respectively each with an extra term to describe the two-way coupling between the shape and the composition of the growth front. This effective description follows both from the reaction-diffusion model and from phenomenological considerations. Therefore, it could apply to other out of equilibrium systems with chiral constituents. For example, the deposition of particles with different chirality and homophilic interactions should be analogous to the growth of cellular aggregates that we studied.
To understand whether chirality can affect fitness, we developed a minimal reaction-diffusion model of growth in dense cellular aggregates such as microbial colonies. This model recapitulated spiralling growth dynamics in colonies composed of cells with the same chirality (Fig. A) and made striking predictions about the competition between strains with different chirality. For strains with the same handedness, but different magnitude of chirality we observed competitive exclusion of the less chiral strain. In contrast, strains with the opposite handedness stably coexisted with the relative abundances of the strains determined by their relative chirality (Fig. B).
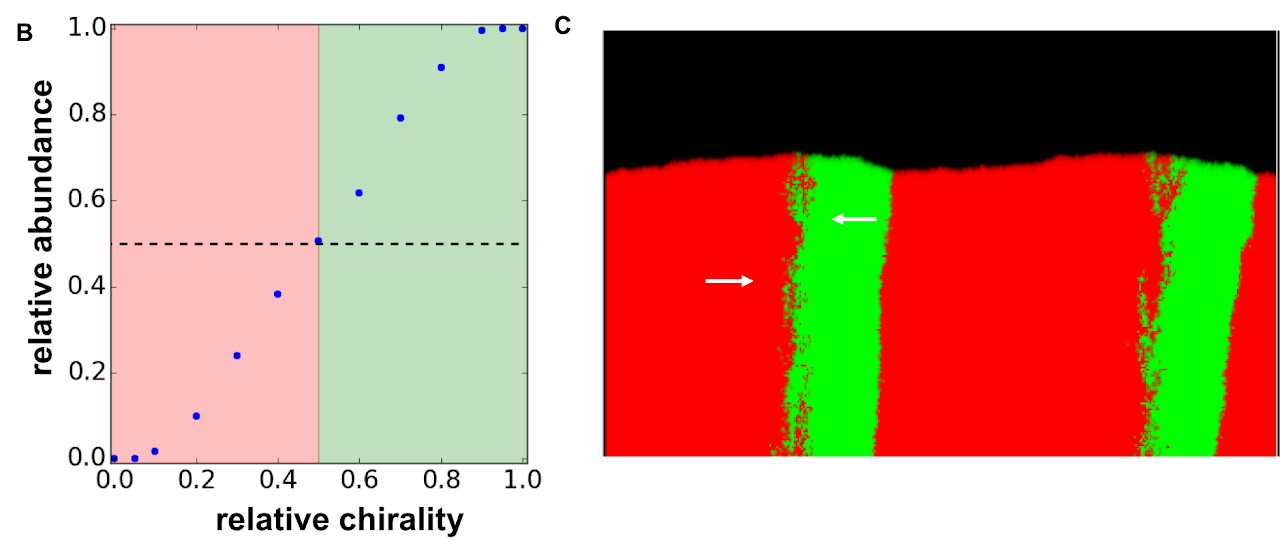
We also found that, under some conditions, the strains with opposite chirality can completely intermix with each. This mixing can greatly facilitate mutualistic interactions such as cross-feeding. One of our future goals is to understand the mixing transitions in more detail and to explore the role of chirality in social interactions between microbes.

For more details see the following paper:
Microtubules are relatively large and rigid biopolymers within the cell that perform numerous functions. For example, microtubules provide mechanical rigidity and act as highways for intracellular transport. Many diseases are caused by microtubule defects, and a lot of effort went into characterizing mechanical and structural properties of microtubule complexes. The assembly of such complexes, however, is a lot less understood even though microtubule complexes are very dynamic and need to reassemble every time the cell moves or divides. In collaboration with Tim Mitchison at Harvard Medical School, we investigated large microtubule complexes called asters. Asters facilitate cell division and are a very convenient experimental system because they take nearly half an hour to assemble and reach about a millimeter in size in early fish and frog embryos.
Traditionally the growth of asters was attributed to the growth of individual microtubules (Fig. A). Each microtubule switches between an elongating and a shrinking state, so depending on the velocities and switching rates a microtubule can either become infinitely long or vanish after a few cycles of growth and shrinkage. Thus, a simple model for aster formation is nucleation at centrosomes followed by elongation of individual microtubules. The resulting structure should become less dense away from the centrosomes, which contradicts experimental observations. Moreover, Mitchison lab found strong evidence of microtubule nucleation away from the centrosomes suggesting that other growth mechanisms are at play.
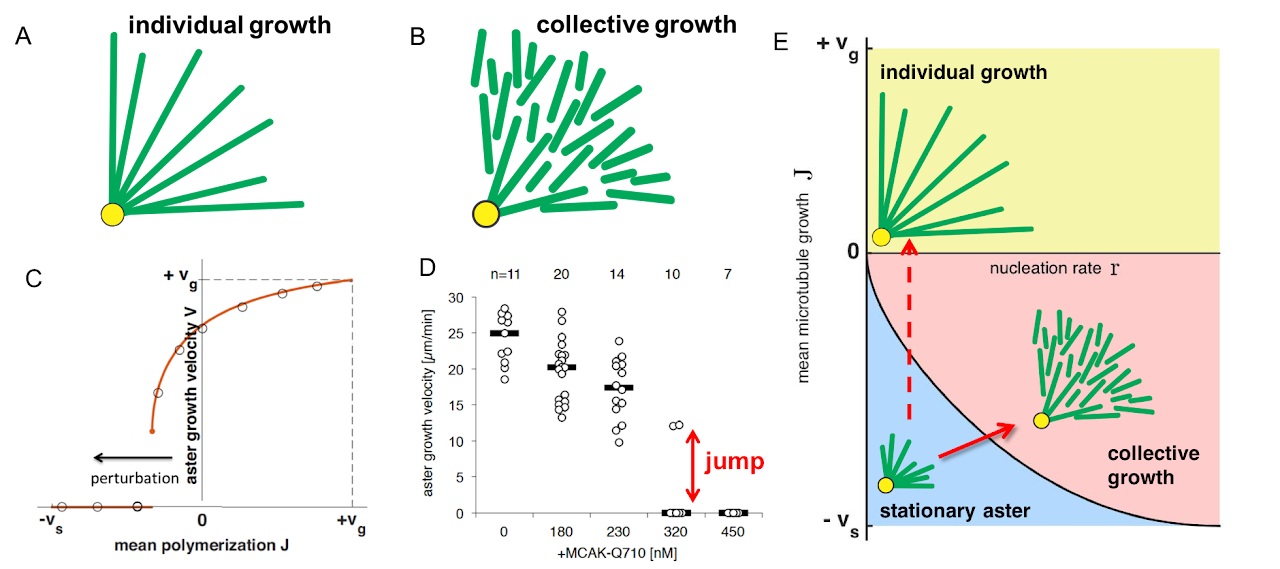
Fig. D: Equations for the effective theory. Colony shape is described by height, h, and colony composition by relative abundance, f. Both h and f depend on time t and position along the growing edge x.
We developed a biophysical model that combines the growth dynamics of individual microtubules with autocatalytic nucleation of new microtubules. In this model, microtubules are shot-lived, but the whole structure grows because each microtubule nucleates on average more than one new microtubule during its lifetime. This model predicts a time invariant density profile that expands at a constant velocity in agreement with experimental observations. The model further explains the self-healing nature of asters and their rapid disassembly at the end of the cell cycle.
One unusual prediction of our model is the jump from zero to nonzero velocity as model parameters are varied (Fig. C). This jump is specific to the structure of our model because jumps occur neither in the traditional model nor in the reaction-diffusion equations typically used to describe spatial growth. We confirmed the existence of this jump by perturbing microtubule dynamics using a mutant version of MKAC protein (Fig. D), which provides further support to the proposed mechanisms of autocatalytic nucleation.
In summary, microtubule complexes could assemble not only via the growth individual microtubules, but also via collective growth with a dynamic balance between nucleation and loss (Fig. E).
For more details see the following paper:
Our intestine is home to hundreds of species that train our immune system, produce essential metabolites, and resist pathogens. This peaceful and often beneficial relationship between us and microbes can break down triggering gut inflammation and microbial dysbiosis. Recent advances in sequencing technologies allowed detailed characterization of the changes in microbial abundances between health and disease. However, translating such data into clinical insights has proven difficult. We recently developed a set of computational tools to address this challenge and used them to understand microbial dysbiosis in Crohn’s disease, a type of inflammatory bowel disease, which affects close to a million people in the USA alone.
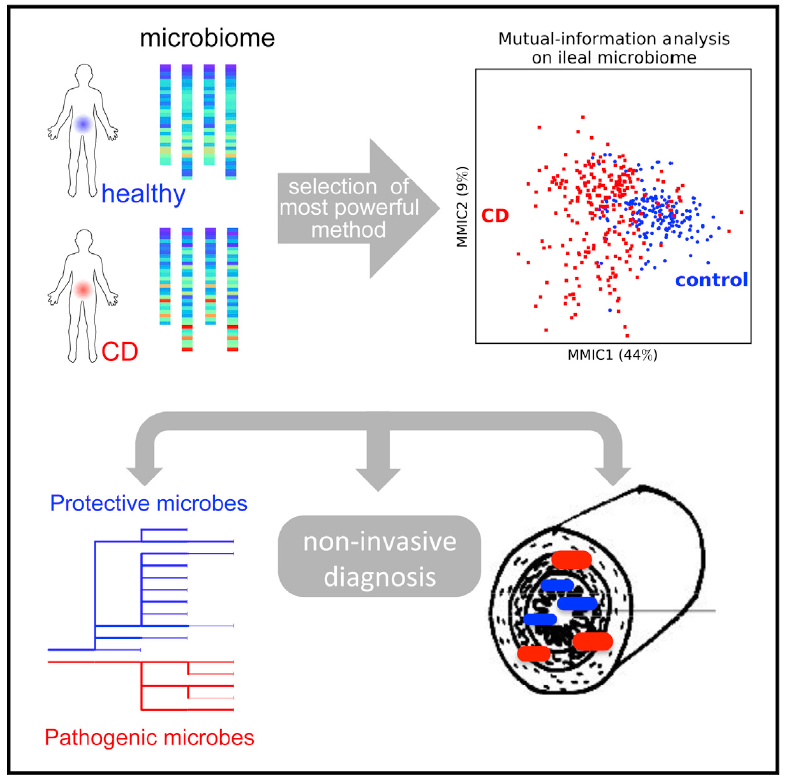
Analysis of microbiome data is often complicated by broad abundance distribution of different taxa and high patient-to-patient variability. We developed a set of statistical techniques to make the patterns of microbiota changes more clear and robust. The power of this approach was demonstrated using two major data sets on pediatric Crohn’s disease. Our method required fewer samples for similar statistical power and identified additional taxa associated with the disease. Discovered associations could distinguish patients with and without Crohn’s disease using a new classifier and depiction tool that identifies directions in the multi-dimensional space of microbial abundances with maximal information on the diagnosis. This maximal mutual information components analysis was superior to the commonly used principle component analysis and remained informative when validated on an independently obtained dataset.
We found that health- and disease-associated bacteria have distinct ecology and evolutionary history. Bacteria correlated and anti-correlated with disease formed two largely non-overlapping phylogenetic trees, suggesting that factors promoting health or disease have deep evolutionary roots and are not frequently exchanged between gut bacteria. Moreover, bacteria that proliferate in Crohn’s disease are preferentially associated with lining of the gut, whereas bacteria decreased in disease reside mostly in the stool. This connection between dysbiosis in lumen and mucosa compartments of the gut enabled patient classification using either biopsies or stool samples with about equal accuracy. Thus, contrary to previous suggestions, stool samples have a high potential for noninvasive diagnostics.
Our study demonstrates that quantitative analysis of microbial dysbiosis can yield insights into disease biology and provide clinically useful predictions.
For more details see our paper:
Feng Wang, Jess L. Kaplan, Benjamin D. Gold, …, Towia A. Libermann, Harland S. Winter, and Kirill S. Korolev, Detecting Microbial Dysbiosis Associated with Pediatric Crohn Disease Despite the High Variability of the Gut Microbiota, Cell Reports 14, 945-955 (2016).
Can one get ahead by staying put? We show that the answer is yes for species colonizing new territories cooperatively.
Population expansions trigger many ecological and biomedical transitions such as the invasion of non-native species or tumor growth. Indeed, spatial spreading is the only mechanism for species to become highly abundant whether we are considering a bacterial colony growing on a Petri dish or the human expansion across the globe. Many invasions are unwelcome because they threaten biodiversity, agriculture, or human health. Controlling such invasions is difficult because evolution often selects for greater dispersal and accelerates the spreading rates. For example, the expansion rate of cane toads in Australia has increased nearly five fold since the invasion began. Invasion acceleration supports the simplest view of evolution as “the survival of the fastest” and provides little hope for management strategies to preserve biodiversity or reduce agricultural losses.
In a recent paper inPRL, we report that invasion acceleration occurs only under somewhat restrictive assumptions, and, in general, evolution can select for both faster and slower dispersal. The direction of evolution depends on cooperation and other forms of positive feedback in population growth. In highly cooperative populations, evolution selects for slower dispersal and eventually arrests further invasion. In contrast, invasions accelerate when cooperation plays a minor role in population dynamics. Thus, “the survival of the fastest” changes to “the survival of the slowest” as population growth becomes more cooperative; see Figure.
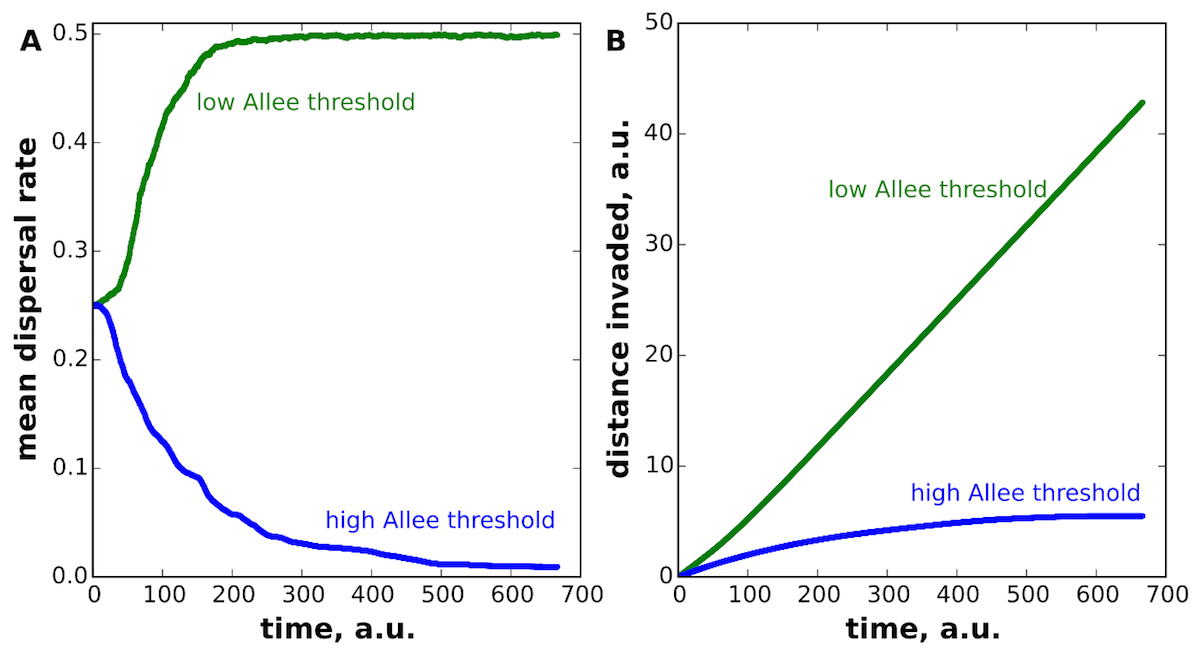
Figure: An illustration of the variability in the gut communities, our new analytical techniques, and major outcomes of the analysis.
Cooperative growth, known as Allee effects ecology, is common in nature because organisms in larger population are more successful at finding mates, defending against predators, and collective feeding. We show that mutations that reduce dispersal have a competitive advantage in cooperative populations even when they have no effect on the growth rate. Dispersal reducing mutations then steadily accumulate at the expansion front bringing invasion to a halt. Invasion deceleration does not require costs of dispersal or fixations of deleterious mutations due to the high genetic drift at the expansion frontier. Instead, our findings are a rare example of evolution deterministically driving the population into unfavorable state and could lead to new strategies to combat unwelcome invaders.
Our results might explain the success of pest-management strategies that make the growth more cooperative by elevating the Allee effect. Even more strikingly, our work suggests that natural selection rather than genetic drift could explain the puzzling loss of the ability to fly by the females of the European gypsy moth, one of the most damaging pests invading North America. These moths have a strong Allee effect because they fail to find mates at low densities, and our theory predicts that reduced dispersal could be favored by natural selection. Flightless females could therefore be an unexpected adaptation to life at the expanding frontier.
Although the evolution in dynamic spatial populations is typically so complex that it can only be studies by numerical simulations, we obtain exact analytical formulae for the selective advantage of mutants with different dispersal rates. Our results show that mutants that disperse either too slowly or too fast compared to the wild type are outcompeted by mutants with intermediate changes in dispersal. Once these intermediate mutants reach fixation they enable the fixation of mutants continue to either increase or decrease the dispersal rates of the population depending on the strength of the Allee effect; see Figure.
For more details see our paper:
Kirill S. Korolev, Evolution arrests invasions of cooperative populations, Physical Review Letters115, 208104 (2015).
Our health, industries, and environment rely on complex microbial communities to provide essential services such as digestion of food or production of biofuels. Microbes cooperate to perform these task by exchanging small, diffusible molecules that affect their growth and function. Evolutionary game theory is the main theoretical framework to study microbial population dynamics, yet it neglects the diffusive nature of microbial interactions. To overcome this limitation, we formulated and solved a model that accurately represents the physics of diffusion in microbial colonies. Specifically, we focused on two-way cross-feeding, a common interaction motif in the microbial world that is the subject of several experimental studies.
Figure: (A) Dispersal increases in cooperative populations, but decreases in non-cooperative populations. (B) As a result, invasions of non-cooperative populations accelerate, but invasions of cooperative populations come to a halt. The transition between these two evolutionary regimes occurs at a moderate value of the Allee threshold (the minimal population density required for growth) relative to the carrying capacity.
Naively one would assume that nutrient diffusion should facilitate mutualistic interactions in microbial colonies. Indeed, because microbes are not completely mixed inside a colony, different species tend to form small domains, and diffusion should facilitate the exchange of the nutrients between the two cross-feeding species. We, however, found that nutrient diffusion reduces the strength of mutualism by promoting spatial demixing, which can trigger complete separation between the species and thus destroy mutualism. When mutualism does persist, species evolve to either increase or decrease the diffusion constants of the nutrients they produce depending on the details of the competition. This observation might explain the large variation in the diffusivities of many microbial secretions that perform essentially the same function.

Figure: Game theory was developed for pairwise interactions, but is commonly applied to microbes with collective interactions mediated by small diffusible molecules.
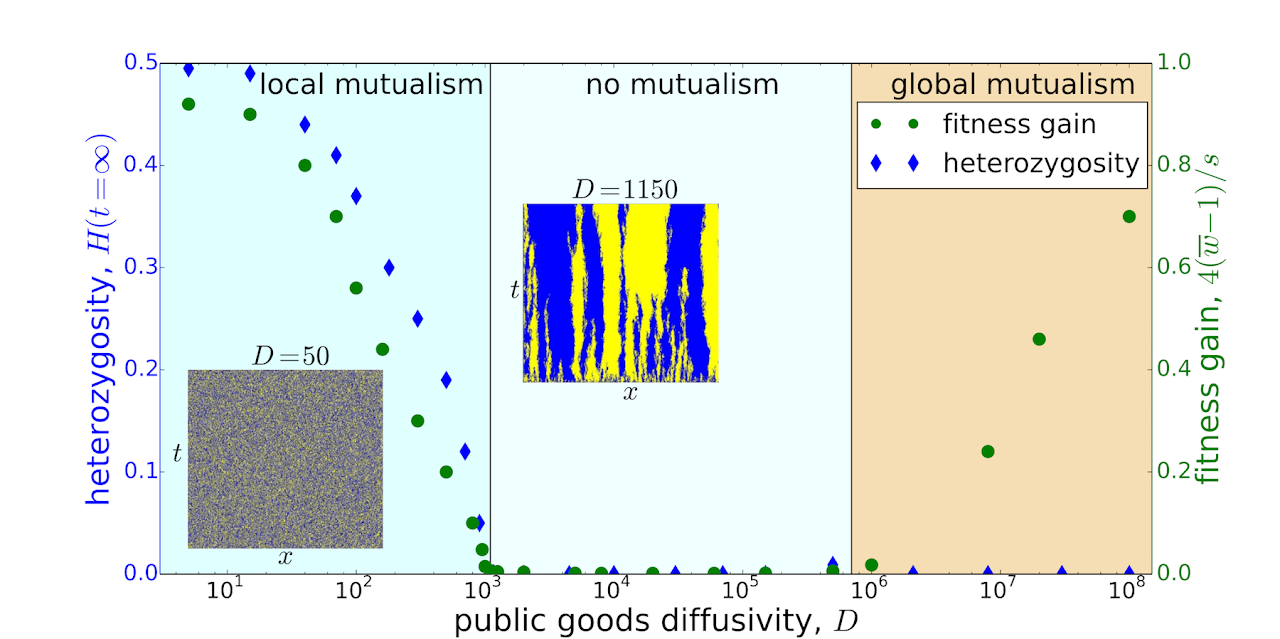
Figure: Local heterozygosity, the measure of species intermixing, decreases with the diffusivity of the public goods, and the species coexistence is completely lost above a critical diffusivity. The fitness gain due to mutualism also decreases and eventually vanishes as the diffusivity increases, unless the diffusion is so fast that nutrients spread through the entire population, and mutualism emerges on a global scale. The two insets show how the distribution of species labeled by different colors varies with position (x-axis) and time (y-axis) for small diffusivities (left) and just below the critical diffusivity (right).
In addition to these specific results, we showed that diffusion-based models are, in fact, equivalent to much simpler models in evolutionary game theory, but with parameters renormalized by diffusion. This observation makes many of the results in game theory applicable to microbes. At the same time, our work shows that the games that microbes play depend on the physical aspects of the environment such as the rate of nutrient diffusion. Thus, one could modify nutrient diffusion to disrupt pathogenic communities or foster beneficial interactions among the microbes.
For more details see our paper:
Rajita Menon and Kirill S. Korolev, Public good diffusion limits microbial mutualism, Physical Review Letters 114, 168102 (2015).
Cancer is an outcome of somatic evolution. To outcompete their benign sisters, cancer cells need to acquire several heritable changes that increase their proliferation. Termed driver alterations, or simply drivers, these heritable changes arise due to a variety of mechanisms, but are often called mutations for simplicity. From the evolutionary point of view, drivers are considered to be beneficial mutations because they provide a selective advantage to the cancer cells. In contrast to other organisms, cancer cells have a lot of room for improvement because normal cells are not optimized for maximal somatic growth. Nonetheless, in the vast space of all possible mutations, the drivers are outnumbered by neutral and deleterious mutations. Recent sequencing studies indeed found thousands of genetic alterations in cancer genomes, despite the fact that only one to ten drivers are thought to be necessary for tumorigenesis. These non-driver mutations are often termed passengers to indicate their irrelevance to cancer progression even though little evidence exists to support this common assumption. On the contrary, our recent analysis of cancer genomes showed that a substantial fraction of passengers in sequenced cancers is likely to be deleterious to the tumor.
Given that mutations are much more likely to be deleterious than beneficial, populations are always in danger of losing viability. Indeed, ‘mutational meltdown’ has been observed in empirical studies of viruses, mitochondria, and bacteria. Cancer tumors are no exception. In very small tumors, slightly damaging passengers can easily spread by chance and beneficial drivers appear rarely. In contrast, natural selection is more efficient at weeding out deleterious passengers in larger tumors, and drivers appear more frequently because more cells can mutate. These dynamics result in a runaway process: small tumors accumulate more damaging mutations, lose fitness and get smaller, whereas large tumors accumulate more drivers, gain fitness and get bigger (see Figure a). Progression to cancer is then akin to overcoming the activation energy of a chemical reaction, because tumors have to rely on the stochasticity of mutation to overcome the critical population size that is necessary for deterministic expansion.
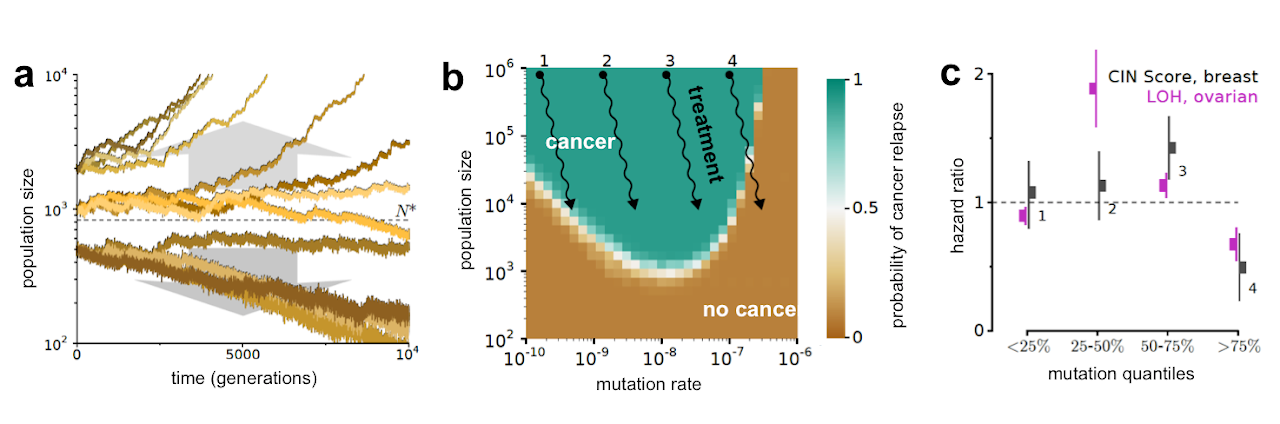
Figure: Damaging passengers create a barrier to cancer progression. (a) The outcome of tumor evolution is bistable: small tumors tend to go extinct via mutational meltdown while larger tumors progress to cancer. The critical population size N∗ necessary for deterministic progression is labeled with a dashed line. (b) Probability of cancer relapse as a function of tumor size and mutation rate following treatment (shown with wavy arrows). Treatments 1 and 4 take patients outside the region where cancer evolution is likely so they are expected to succeed. In contrast, treatments 2 and 3 are expected to fail due to cancer evolution. (c) Consistent with this prediction, the hazard ratio, i.e. relative death rate, peaks at intermediate rates of a mutational process (chromosomal aberrations or loss of heterozygosity). The data are taken from Birkbak, N. J. et al. Cancer research 71, 3447 (2011) and Wang, Z. C. et al. Clinical Cancer Research 18, 5806 (2012).
The critical population size, which controls the ability of a tumor to adapt, depends on the mutation rate and fitness cost of deleterious passengers. The dependence on the mutation rate is illustrated in Figure b. As expected, cancer evolves slowly at low mutation rates because driver mutations are rare. However, cancer progression is also inhibited at high mutation rates primarily due to the little-known, but important phenomena of passenger mutations interfering with the fixation of driver mutations. This effect of the mutation rate on the tumor’s evolutionary potential has been repeatedly observed in clinical studies (Figure c) supporting the idea that damaging passengers play an important role in cancer progression.
Our work may have direct clinical implications because treatments can in principle modify evolutionary parameters. Mutation rates can be increased by adding a mutagen or by inhibiting DNA repair machinery. Likewise, the fitness costs of passengers can be increased by inhibiting the cellular machinery that buffers against perturbations. In fact, such treatments are already used in the clinic, although motivated by different mechanisms of action. For example, proteasome inhibitors have been approved for the treatment of multiple myeloma, and chaperone and PAPR inhibitors are in clinical trials. Moreover, proteins with passenger mutations can be recognized as foreign and trigger an immune attack; thus, the success of immunotherapies to a large extent depends on the genetic load accumulated by the tumor.
Species could become endangered due to a variety of factors including climate change, pollution, habitat destruction, or overfishing. Forecasting and potentially averting species extinction is important not only for the preservation of biodiversity but also for economic reasons since agriculture and fishing depends on healthy ecosystems and abundant stock. Quite surprisingly, it can be difficult to see if a species is endangered simply from its total abundance because population dynamics in ecosystems is often very nonlinear, and many species grow poorly at low densities (Allee effect). As a result, many populations collapse if their density falls below a critical threshold. Although the vicinity of such a threshold is not obvious from the population size alone, fluctuations of the population size and its spatial variation can in principle be used to anticipate the approach of the tipping point. In collaboration with Lei Dai and Jeff Gore, we showed that this is indeed possible using an experimental yeast population. We also found that signals based on fluctuations are weaker in spatial populations, although that is compensated by a deterministic indicator of ecological resilience: recovery length. Recovery length is a typical distance over which spatial perturbations decay, and it increases sharply before the tipping point. This behavior is analogous to the divergence of the penetration depth in superconductors near the phase transition.
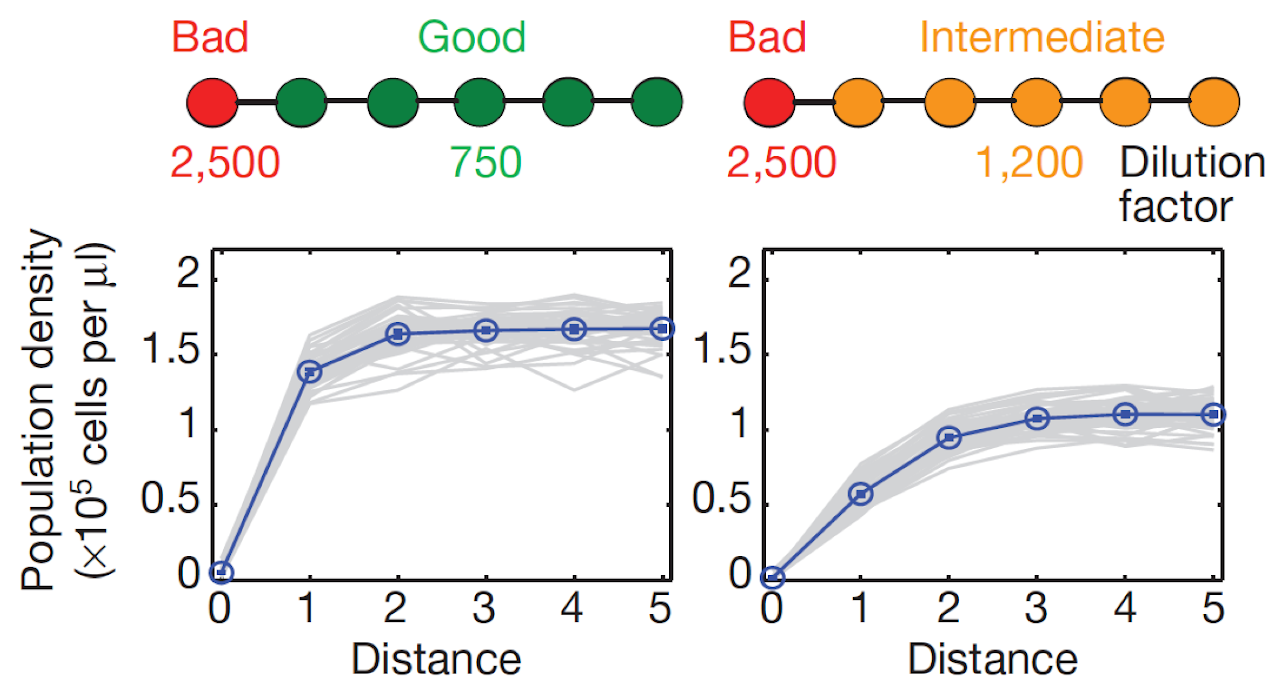
Figure: This data is from an experiment in a microbial population. The left most patch is of very low quality, and the population density there is very small. The population density increases to the right as the environmental quality improves over a characteristic length: the recovery length. When the environment on the right is far from a tipping point, the recovery length is short, but, near the transition, the recovery length becomes very large.
Species expand their geographical ranges following an environmental change, long range dispersal, or a new adaptation. Range expansions not only bring an ecological change, but also affect the evolution of the expanding species. One major effects of range expansion is an increased role of genetic drift, i.e. stochastic changes in the frequencies of different genotypes. However, even when stochastic effects are negligible, range expansion can profoundly alter the course of evolution provided evolutionary and ecological dynamics are coupled. Interestingly, range expansion can increase the level of cooperation in the population.
Cooperative traits like the production of a public good are often under frequency-dependent selection, such that defectors (also known an cheaters or non-producers) outcompete cooperators when the latter are in large numbers and public goods are abundant, but cooperators prevail when they are rare and public goods are scarce. Although often neglected by evolutionary models, frequency-dependent selection is affected by the population size. In particular, smaller populations maybe more permissive of cooperation than larger populations. The population size is in turn affected by the overall fitness of the population, which depends on the number of cooperators.
When population expand to new territories, this eco-evolutionary feedback increases the number of cooperators at the expansion edge and, under certain conditions, allows them to escape and outrun defectors. Following the separation, cooperators colonize the new territories while defectors slowly invade them from behind. In populations where range expansions occur frequently (e.g. due to forest fires), cooperation could be maintained at high levels by this mechanism of spatial escape of cooperators.
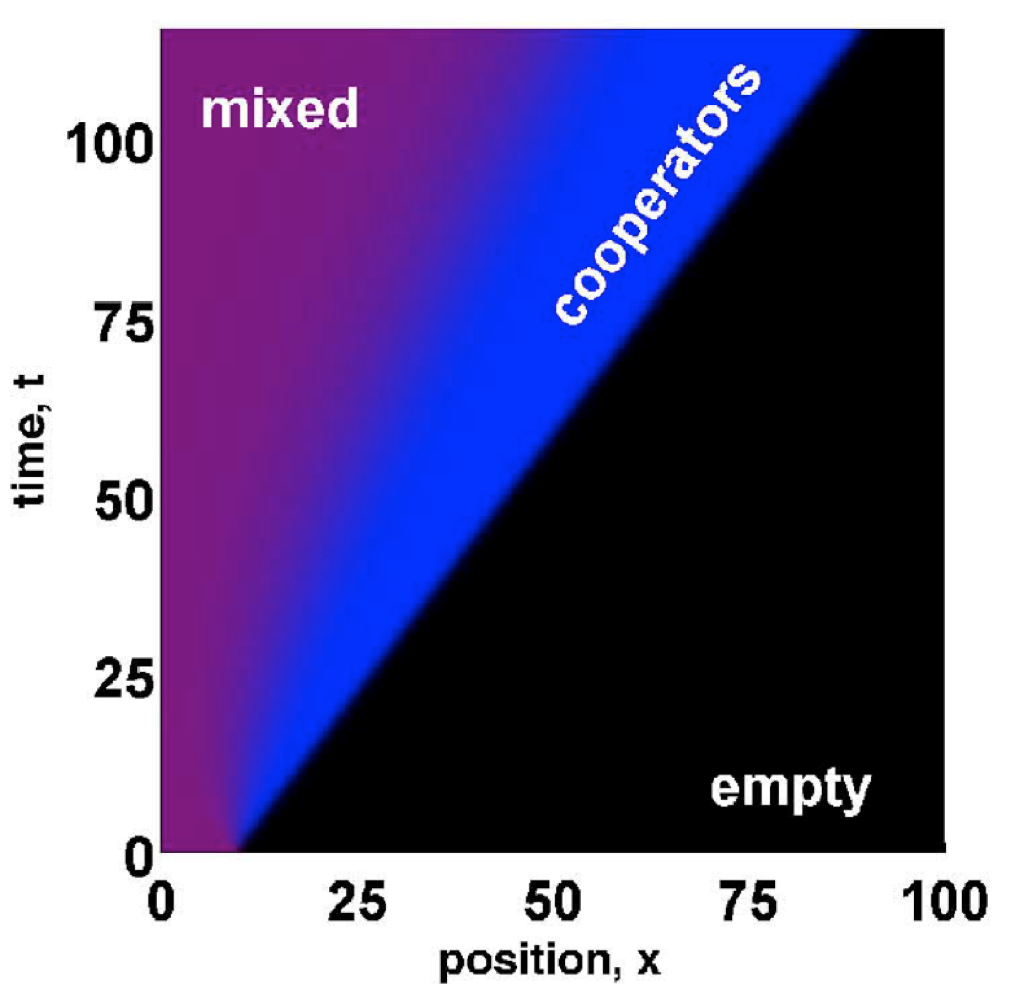
The evolution and ecology of cooperation as well as other social traits can be studied in microbial populations that reproduce rapidly and are easy to manipulate. In collaboration with Manoshi Datta and Jeff Gore, I studied range expansions of cooperators using an experimental yeast population. Yeast cells naturally cooperate to digest sucrose. Digestion occurs outside the cell, which allows defectors to benefit from the digestion products without investing into the enzyme necessary for sucrose breakdown. Using this system, we indeed found that cooperators are favored at the front of the expansion and can outrun defectors.
Cooperation is at the heart of many complex systems. On an organism level, gut bacteria help their hosts digest cellulose. On an ecosystem level, plants often rely on fungi to receive important nutrients. Even human societies are products of cooperation between individuals. Despite the apparent advantage and pervasiveness of mutualistic interactions, their existence is often difficult to explain by a naive application of Darwinian natural selection. Cooperation is known to be susceptible to cheating; surprisingly, cooperation can also be destroyed by number fluctuations.
Figure: This plot shows that, under some conditions, the range expansion from an initially well-mixed population can split into two population waves. The first wave (blue) is the expansion of cooperators, which dominate the frontier. The second wave is the invasion of defectors (red) into the region already colonized by cooperators resulting in a mixed population (magenta). Note that these two waves move with distinct velocitiesIn collaboration with David Nelson from Harvard University, I modeled mutualistic interaction in spatially extended populations using evolutionary game theory. In addition to cooperation, we modeled migration of organisms and number fluctuations due to discreteness of the number of organisms and the stochastic nature of reproduction and death. Fluctuations and migration are important because they lead to genetic demixing, i.e. spatial segregation of different genotypes in space. Since mutualism is only possible when different species interact locally, genetic demixing should inhibit cooperation. Indeed, we found that, even when mutualism confers a distinct selective advantage, it persists only in populations with sufficiently high density and frequent migrations. When these parameters are reduced, mutualism is lost through a nonequilibrium phase transition. Nonequilibrium phase transitions share many features with their equilibrium counterparts; in particular, they can often be assigned to universality classes. We found that mutualism is generically lost through a second order phase transition in the universality class of directed percolation (DP), except when the two species benefit equally from the interaction. In this symmetric case, the transition belongs to the DP2 universality class. The phase diagram is strongly influenced by the crossover form DP2 to DP transition, similar to the bicritical point in equilibrium statistical mechanics.
The evolution of natural populations involves more than mutations followed by natural selection: Reshuffling of genes during genetic recombination, stochasticity, and spatial migrations are also important. When organisms spread to new territories, the effects of fluctuations and spatial structure become especially pronounced because the number of organisms at the front of the expansion is typically small. Such expansions are common in nature, a disease outbreak being a familiar example, yet remain poorly understood. Recently, we developed several mathematical models and quantitative experiments to illuminate the effects of demographic stochasticity and natural selection on the evolutionary dynamics of expanding populations.
Essential features of range expansions can be captured by the approximation of “dimensional reduction”: Instead of modeling the two-dimensional growth of the population, one considers only the dynamics of a quasi one-dimensional population near the expansion front. In this simplified picture, the relatedness between any two randomly sampled organisms can be easily calculated. We found that the randomness of reproduction combined with the quasi one-dimensional spatial structure leads to a significant loss of genetic diversity. Moreover, the expansion creates a sectoring spatial structure behind the front, see Fig. 2. This structure strongly affects other evolutionary processes; for example, we found that selective sweeps occur more slowly in expanding populations.
Figure: Mutualism in spatial populations. Three sets of simulations are shown with increasing selective advantage of mutualism s. For weak mutualism, the two species demix and the cooperation is lost. For strong mutualism, the mixed, cooperative state persists.
We used microbial colonies to test the predictions of our theoretical work on range expansions. Microbes are particularly suitable for this purpose because recent advances in molecular microbiology and modern genetics enable great control over the behavior of cells. Moreover, the small size and short generation times make it possible to observe the dynamics on ecological and even evolutionary time scales. In collaboration with the experimental groups of Andrew Murray and Kevin Foster, we developed a set of methods to estimate important biological parameters of microbial colonies (migration rate, relative fitness, and effective population size responsible for the number fluctuations) from the observed spatial patterns like the one shown in the Figure above.
The rate at which a new advantageous mutation takes over the population is an important quantity because it sets the time scale of evolutionary change. In a spatially extended habitat, a beneficial mutation creates a genetic wave expanding from the location where the mutation occurred. The spreading of beneficial mutations or infectious diseases can usually be tackled when randomness is small, but chance often plays a major role in nature. In collaboration with Oskar Hallatschek, I studied how an advantageous mutation spreads in the limit of strong fluctuations. Mathematically, this spreading is described by the stochastic Fisher-Kolmogorov-Petrovsky-Piscounov equation, a classical model in nonequilibrium physics, which is also used in chemical kinetics, ecology, and nuclear physics. We developed a powerful technique to analyze systems in which the effects of chance dominate deterministic forces and calculated the expansion velocity in one and two spatial dimensions. We also analyzed the population structure in the hybrid zone, where both the mutants and the wild type are present. Instead of a stationary, smooth transition region predicted by the classical no-noise approximation, we found non-trivial dynamics of a few rugged kinks that diffuse, give birth by division, and annihilate upon encounter.
Emulsions are a very useful state of matter with applications ranging from paint manufacturing to drug delivery. Even cellular membranes are hypothesized to be emulsions of lipid rafts—islands of saturated lipids—floating in the sea of unsaturated lipids. The physical properties of emulsions strongly depend on the interactions among the embedded droplets. In fact, metastable emulsions can persist only if the coalescence of inclusions is arrested by the short-range repulsion of the inclusions. Usually, this repulsive force arises due to Coulomb or steric interactions. With my PhD advisor, David R. Nelson, I studied a novel stabilizing mechanism in topological emulsions, that is emulsions where the continuous liquid phase has an orientational (liquid-crystal-like) order parameter.
In topological emulsions, the interactions among the inclusions arise due to anchoring boundary condition on the surface of the droplets and elastic energy associated with the deformation of the continuous ordered phase. Furthermore, these interactions are mediated by topological defects (singularities in the texture of the liquid crystal). We found that two droplets attract at long separation, but repel at short separations, which results in a well-defined separation between the inclusions and stabilization of the emulsion. Interestingly, this formation of dimers resembles ordinary chemical bonds, but with topological defects instead of electrons, and leads to self-organization of the inclusions.
High-temperature superconductors play an important role in modern technology, for example, as sources of strong magnetic fields. Although superconductors have vanishing resistivity, high-temperature superconductors dissipate energy when the magnetic field is strong enough to form vorticies in the bulk of the material. This dissipation is caused by the frictional forces during the motion of the vorticies as they jump from one defect in the material to another under the influence of the Lorentz force. As a result, the magnetization of a premagnetized superconductor decreases with time. With my research advisor, L.S. Uspenskaya, I studied how the rate of this decrease depends on the spatial configuration of the magnetic flux and the geometry of the magnet. We found that for thin samples the relationship between the super-current and magnetic field becomes very nonlocal, which leads to inhomogeneous magnetization relaxation in the sample. Consequently, a judicious choice of the spatial pattern of magnetization may lead to slower relaxation.
Figure: Spatial segregation during an outward expansion of a Pseudomonas aeruginosa colony seen in a fluorescent microscope. The Petri dish was inoculated with a well-mixed population of two equally fit strains that differ only in the color of fluorescent markers. As this population expands, it segregates into well-defined domains. The colony is of the order 2 cm in radius.